ASTRE & CUTASTRE : (Accelerated) A contrario Smooth TRajectory Extraction
Table of Contents
1 Description
Our work started with this simple observation: the human visual system is able to perceive motion hidden in high amounts of noise. How far could a computer go?
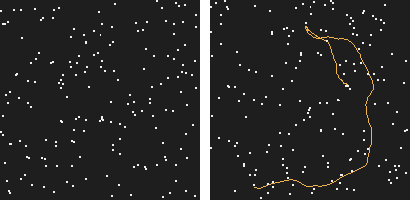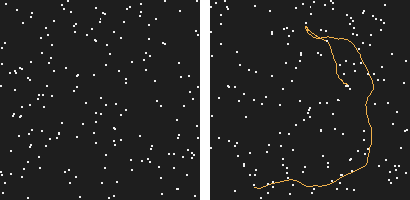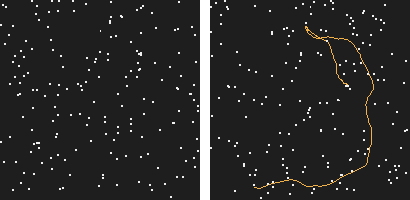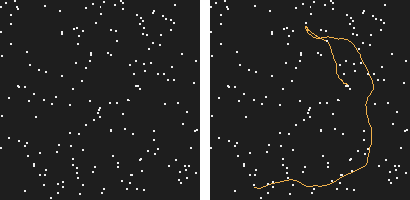
Figure 1: This is a 200 frames sequence of 180 noise points and one real trajectory. Can you spot it?
M. Primet and L. Moisan first proposed the ASTRE (A-contrario smooth trajectory extraction) framework based on the a-contrario methodology that defines a (quasi-parameterless) perceptual metric on the trajectories, and an algorithm based on dynamic programming to extract the trajectory having the best appearance.
The principle of a-contrario algorithms is to control the number of detections that are made in pure noise (that is false detection), and our method is thus resilient to high amounts of noise. The resulting perceptual metric can also be used to filter the result of any other tracking algorithm, hence reducing the number of false detections.
If the detection of smooth trajectories in a noisy point set sequence can be realized optimally with the ASTRE algorithm, its quadratic time and memory complexity with respect to the number of frames is prohibitive for many practical applications. R. Abergel and L. Moisan proposed a variant named CUTASTRE which results in a linear complexity with respect to the number of frames, without affecting in practice the detection performances.
2 Scientific papers and supplementary material
2.1 Papers
- ASTRE : ``Point tracking: an a-contrario approach'' (Maël Primet, Lionel Moisan), preprint
MAP5 nº2012-06 (download preprint MAP5

 ).
).
- CUTASTRE : ``Accelerated A-contrario Detection of Smooth
Trajectories'' (Rémy Abergel, Lionel Moisan), proceedings of the European
Signal Processing Conference (Eusipco), 2014 (download published version
 ,
,  , revised preprint
, revised preprint
 ).
).
2.2 BibTeX citations
@article{astre2012,
author = {Primet, Ma\"{e}l and Moisan, Lionel},
title = {Point tracking: an a-contrario approach},
year={2012},
}
@inproceedings{cutastre2014,
author = {Abergel, R\'{e}my and Moisan, Lionel},
title = {Accelerated A-contrario Detection of Smooth Trajectories},
booktitle = {Proceedings of the European Signal Processing Conference (Eusipco)},
year = {2014},
}
3 Download the code
3.1 License
This program is released under GPL License.
Feel free to use and adapt this code. However, if you use it for your research or in software code, please be so kind as to cite the papers [1,2].
Also, if you use, adapt or improve the code, we would love to hear about it!
4 Installation
4.1 Content and dependencies
This software is designed to have very low dependencies. Only the following standard libraries are required :
stdiostdlibstringmath
It is composed of three independent source files astre_noholes.c and
cutastre_noholes.c, corresponding to ASTRE and CUTASTRE implementations,
and metrics.c, a tool dedicated to the evaluation of the detection
quality by mean of comparison of a given sequence with a ground truth
sequence. Those files must be compiled separately.
4.2 Compilation with gcc
Open a terminal, then from the src directory of this software, run the gcc
command lines displayed below (of course this can be adapted for others
C-compilers).
gcc -O3 astre_noholes.c -lm -fopenmp -o astre_noholes gcc -O3 cutastre_noholes.c -lm -fopenmp -o cutastre_noholes gcc -O3 metrics.c -lm -o metrics
The executable files astre_noholes, cutastre_noholes and metrics are
then created into the same repertory.
4.3 Check the installation
To check the installation was correctly done, you run ASTRE and CUTASTRE on a sample point set sequence (bundled with this software).
First, let us test the astre_noholes program. From the src directory,
execute
./astre_noholes -v ../data/snowseq.txt /tmp/out_astre.txt
This must result in the extraction of 39 trajectories (go here to visualize the terminal standard output).
Now let us test the cutastre_noholes program:
./cutastre_noholes -v ../data/snowseq.txt /tmp/out_cutastre.txt
This must result in the extraction of 38 trajectories (go here to visualize the terminal standard output).
Last we check the metrics tool:
./metrics ../data/snowseq.txt /tmp/out_cutastre.txt
Go here to visualize the terminal standard output.
5 Tutorial
5.1 Usage of ASTRE & CUTASTRE
As usually, optional inputs are displayed inside square brackets.
Usage of ASTRE.
Usage: astre_noholes [--help] [--version] [-W width] [-H height] [-S maxspeed]
[-m minscore] [-T numthreads] [-v] filenamein filenameout
--help : display help
--version : display program version
-W width : set the width (in pixels) of the image domain
(default = Xmax-Xmin+1, X(min/max) = smallest/highest
X-coordinate (horizontal axis) of the input sequence)
-H height : set the height (in pixels) of the image domain
(default = Ymax-Ymin+1, Y(min/max) = smallest/highest
Y-coordinate (vertical axis) of the input sequence)
-S maxspeed : only detect trajectories with maximal speed (that is
the maximal distance (in pixels) between two consecutive
points) less than maxpseed (may drastically reduce the
execution time)
-m minscore : (default = 0) set the score threshold, only trajectories
with a score higher than minscore will be extracted
-T numthreads : (default = 1) set the number of OpenMP threads
-v : verbose mode
filenamein : path to the input point set sequence file
filenameout : path to the output point set sequence file
Usage of CUTASTRE.
Usage: cutastre_noholes [--help] [--version] [-W width] [-H height] [-S maxspeed]
[-c chunksize] [-o overlapsize] [-m minscore] [-T numthreads] [-v]
filenamein filenameout
--help : display help
--version : display program version
-W width : set the width (in pixels) of the image domain
(default = Xmax-Xmin+1, X(min/max) = smallest/highest
X-coordinate (horizontal axis) of the input sequence)
-H height : set the height (in pixels) of the image domain
(default = Ymax-Ymin+1, Y(min/max) = smallest/highest
Y-coordinate (vertical axis) of the input sequence)
-S maxspeed : only detect trajectories with maximal speed (that is
the maximal distance (in pixels) between two consecutive
points) less than maxpseed (may drastically reduce the
execution time)
-c chunksize : split the frames of the sequence into chunks of specified
size (in number of frames), must be >=3 (default = 20)
-o overlapsize : set the overlap size (in number of frames) between two
consecutive chunks, must be >= 2 and < c (default =
maximal value between 2 and floor(c/2))
-m minscore : (default = 0) set the score threshold, only trajectories
with a score higher than minscore will be extracted
-T numthreads : (default = 1) set the number of OpenMP threads
-v : verbose mode
filenamein : path to the input point set sequence file
filenameout : path to the output point set sequence file
Additional comments about the options
-S maxspeed : the speed threshold maxspeed is a physical parameter that
can be easily adjusted in many applications. The use of this additional
knowledge restricts the number of linking possibilities among the sequence,
and reduces very significantly the execution time in general for both
algorithms. However you must keep in mind that even using a speed threshold,
the complexity of ASTRE remains quadratic with respect to the number of frames
which is too much to handle long data sequences. The long data sequences must
be processed with CUTASTRE which has a linear complexity with respect to the
number of frames.
-c chunksize (CUTASTRE only):
-o overlapsize (CUTASTRE only):
-m minscore : you can consider that the higher is the score of a trajectory,
the better is the confidence related to its detection. More rigorously,
minscore controls the number of false detection that we accept when we
process the sequence, we mathematically proved that in pure noise point set
sequences, the average number of detected trajectories (i.e. the number of
false detections) is less than \(10^{-\text{minscore}}\) (we refer to
[1,2] as well as to the a-contrario methodology for more
complete information). You can remember that we usually set minscore to zero
in order to ensure that (in average) less than one detection is made in random
(pure noise) sequences.
-T numthreads : some parts of the computation have been parallelized with
OpenMP, if you are using a multicore or multi-CPU architecture, you may
reduce a bit the computation time by increasing the number of threads,
unfortunaltely the improvement is not very significant.
5.2 Input/output files and their visualization
5.2.1 Input/output files format
Inputs
The input point set sequences are simple text files where each line
describes a point of the sequence. All lines must start with
3 integers named <frameid>, <x> and <y> separed with spaces
(at least one space between two integers), excepting those beginning
with character # that will not be interpreted (those lines are
comments).
<frameid>(first entry) : identifier of the frame containing the point (frames are numeroted from 0),<x>(second entry) : X-coordinate (horizontal axis) of the point,<y>(third entry) : Y-coordinate (vertical axis) of the point.
Any content that may be present after the third entry <y> of each line
of the input file will be ignored.
Notice that we do not impose any particular order for the lines, more precisely any permutation of the lines of a point set sequence will not change the data set.
Outputs
5.2.2 Sample data
Our software comes bundled with sample data into the data directory. You
can have a look to their content directly in the data directory or by
clicking the links below.
- snowseq.txt : a real-world sequence of falling snow flakes in front of a tainted background. This sequence is presented here in more details.
- psmgshort.txt : a synthetic sequence of 100 frames, containing 20 synthetic trajectories with length between 50 and 100 frames (generated with the Point-Set Motion Generation algorithm (PSMG)) and 10 spurious points per frame.
- psmglong.txt : a longer PSMG sequence of 5000 frames, containing 500 trajectories with length between 100 and 200 frames and 10 spurious points per frame.
Notice that each point of those sequences is described by four entries, the
fourth entry corresponds to the (usually unknown) <trajid> entry (recall that
all points with same <trajid> belong to the same trajectory, points with
<trajid> = -1 are spurious points). Those sequences are actually ground-truths
and can be used as oracles to evaluate the quality of the detection performed by any
algorithm, using for instance the metrics proposed in section 5.4.
When using those three sequences as input for ASTRE or CUTASTRE algorithms, the
<trajid> entry is of course ignored (recall that any content following the third
entry of an input sequence is ignored), as it is precisely the role of the algorithms
to fill this entry for each point of the processed sequence.
5.2.3 Visualization tool [optional]
We provide with our software a trajectory viewer, of course such a graphical
tool fatally involves dependencies, thus in order to use it you must first
install Python and PyQt4. People working with a Debian GNU/Linux
distribution can install those softwares using apt-get.
sudo apt-get install python python-qt4-dev
To visualize a point set sequence, for instance the snow-sequence, from the
src directory of our software (but also from anywhere else by changing the
relative path), you can execute
./vision/tview.sh -t ../data/snowseq.txt
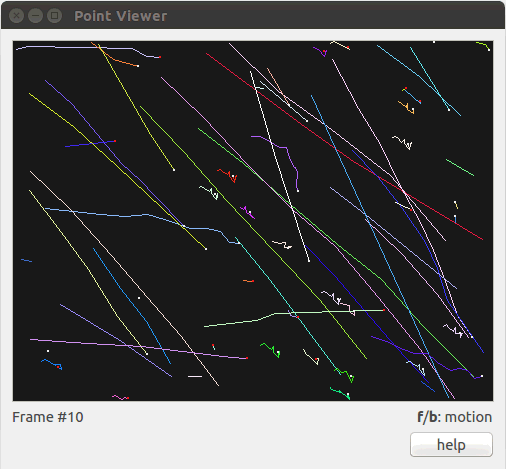
Figure 2: Visualization of the snow sequence using tview. The user uses the keyboard keys f (forward) and b (backward) to move through the sequence. Other interactive keys are availables (zoom in/out, change colors, …), one can click the help button or also use the --help (or -h) option to get more information.
5.3 Examples
5.3.1 Processing the snow sequence
Open a terminal, then from the src directory of our software, run the command
lines displayed below.
First let us run ASTRE and CUTASTRE using default settings and verbose mode
./astre_noholes -v ../data/snowseq.txt /tmp/out1_astre.txt ./cutastre_noholes -v ../data/snowseq.txt /tmp/out1_cutastre.txt
If you have installed our visualization tool tview, use it to
display the outputs
./vision/tview.sh -t /tmp/out1_astre.txt ./vision/tview.sh -t /tmp/out1_cutastre.txt
Now let us use a speed threshold of 200 pixel/frame (with the option -S 200),
this reduces the execution time (for both algorihms).
./astre_noholes -v -S 200 ../data/snowseq.txt /tmp/out2_astre.txt ./cutastre_noholes -v -S 200 ../data/snowseq.txt /tmp/out2_cutastre.txt
Note that the actual maximal speed observed in the snow sequence is less than 161 pixel/frame. When in practice the typical speed of the points of the sequence is unknown, even the use of a pessimistic speed threshold improves the execution time.
You can if you want, change the setting of ASTRE and CUTASTRE parameters. For
instance let us change the setting of the chunksize parameter of CUTASTRE.
Recall that the less is chunksize, the faster is the algorithm, however
keep in mind that it must be chosen high enough to capture the
motion coherence. The setting proposed below is somehow optimal for the snow
sequence.
./cutastre_noholes -c 8 -v -S 200 ../data/snowseq.txt /tmp/out3_cutastre.txt
5.3.2 Processing synthetic sequences
Open a terminal, then from the src directory of our software, run the command
lines displayed below.
First let us run ASTRE and CUTASTRE on the short PSGM sequence (100 frames) using default settings and verbose mode
./astre_noholes -v ../data/psmgshort.txt /tmp/out4_astre.txt ./cutastre_noholes -v ../data/psmgshort.txt /tmp/out4_cutastre.txt
If you have installed our visualization tool tview, use it to
display the outputs
./vision/tview.sh -t /tmp/out4_astre.txt ./vision/tview.sh -t /tmp/out4_cutastre.txt
Now let us consider the long PSMG sequence (5000 frames). Due to the quadratic complexity of ASTRE with respect to the number of frames (both in terms of memory and execution time) this sequence cannot be processed with ASTRE with a standard computer (even using a speed threshold). Thanks to the linear complexity of CUTASTRE with respect to the number of frames, the processing of this sequence is now possible.
./cutastre_noholes -v ../data/psmglong.txt /tmp/out5_cutastre.txt
Note also the nice reduction of the execution time allowed by the use of a speed threshold of 150 pixel/frame (all the more so this choice is again quite pessimistic as the actual maximal speed observed in this sequence is less than 28 pixel/frame).
./cutastre_noholes -v -S 150 ../data/psmglong.txt /tmp/out6_cutastre.txt
5.4 Metrics for the evaluation of the results
A qualitative visual inspection of the found trajectories already gives a rough idea of the algorithms performances. Yet one would sometimes rather have a way to quantify those performances.
Given a reference (or ground truth) sequence and a tested sequence (typically the output detection sequence obtained by running ASTRE or CUTASTRE on the reference), the precision and recall measurements can be used to compare the tested sequence with the reference sequence and evaluate the quality of the detection. Those quantities are defined as
$$ \text{precision} = \frac{\text{number of correct links}}{\text{number of found links}}, \quad\text{recall} = \frac{\text{number of correct links}}{\text{number of real links}},$$
where we call link two consecutive points of a trajectory, possibly separated by a hole (meaning that the two points may not belong to two consecutive frames), and a link is said to be real if it belongs to the ground truth, found if it belongs to the tested sequence, and correct if it belongs to both sequences.
More intuitively, the quantity \(1-\text{precision}\) measures the proportion of false positive links among found links while \(1-\text{recall}\) measures the proportion of false negative links among real links.
Sometimes the precision and recall measurements are combined leading to other quality metrics, such as the \(\text{F}_1\text{-score}\) (harmonic mean between precision and recall) or the G-measure (geometric mean between precision and recall),
$$\text{F}_1\text{-score} = 2 \times \frac{\text{precision} \times \text{ recall}}{\text{precision}+\text{recall}}, \quad \text{G-measure} = \sqrt{ \text{precision} \times \text{recall}}\;.$$
Usage of the metrics tool.
Usage: metrics [--help] [--version] ref seq --help : display help --version : display program version ref : path to the reference point set sequence file seq : path to the tested point set sequence file
Example. Process the (short) PSMG sequence using cutastre and evaluate the
quality of the detection. From the src directory, execute the following
commands
./cutastre_noholes -c 30 -S 100 ../data/psmgshort.txt /tmp/out_cutastre.txt ./metrics ../data/psmgshort.txt /tmp/out_cutastre.txt
6 Contact
This work has been done by Lionel Moisan, Maël Primet and Rémy Abergel.
Université Paris Descartes, laboratoire MAP5 (CNRS UMR 8145), 45 rue des Saints-Pères, 75006 Paris – FRANCE.
If you use, adapt or improve it, we would love to hear about it!

